Example (1D): Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 8: | Line 8: | ||
pdbSetDouble Silicon Test Rel.Error 1.0e-2 | pdbSetDouble Silicon Test Rel.Error 1.0e-2 | ||
Establish the grid and assign a material | |||
line x loc=0.0 spac=0.0002 tag=Top | line x loc=0.0 spac=0.0002 tag=Top | ||
line x loc=0.2 spac=0.001 tag=Bottom | line x loc=0.2 spac=0.001 tag=Bottom | ||
region silicon xlo=Top xhi=Bottom | region silicon xlo=Top xhi=Bottom | ||
init | init | ||
Gaussian implant profile | |||
#fill in file name for as-implanted profile here | #fill in file name for as-implanted profile here | ||
sel z=1.0e20*exp(-(x/0.001)*(x/0.001))+1.0e12 name=Test | sel z=1.0e20*exp(-(x/0.001)*(x/0.001))+1.0e12 name=Test | ||
| Line 21: | Line 21: | ||
sel z=log10(Test) | sel z=log10(Test) | ||
plot.1d max=0.06 min=[list 0.0 12.0] label=implant | plot.1d max=0.06 min=[list 0.0 12.0] label=implant | ||
#value for diffusivity goes here | #value for diffusivity goes here | ||
| Line 29: | Line 28: | ||
term name = TestActive add silicon eqn = "(Test>1.0e20)?1.0e20:Test" | term name = TestActive add silicon eqn = "(Test>1.0e20)?1.0e20:Test" | ||
Establish the physics | |||
#simple Fick's law based diffusion equation | #simple Fick's law based diffusion equation | ||
pdbSetString Silicon Test Equation "ddt(Test) - TestDiff * grad(TestActive)" | pdbSetString Silicon Test Equation "ddt(Test) - TestDiff * grad(TestActive)" | ||
Use "difuse" command to solve | |||
diffuse time = 60.0 temp=550 init=1.0e-1 | diffuse time = 60.0 temp=550 init=1.0e-1 | ||
| Line 50: | Line 47: | ||
sel z=log10(model) | sel z=log10(model) | ||
plot.1d !cle max=0.06 min=0.0 label=analytical_diff | plot.1d !cle max=0.06 min=0.0 label=analytical_diff | ||
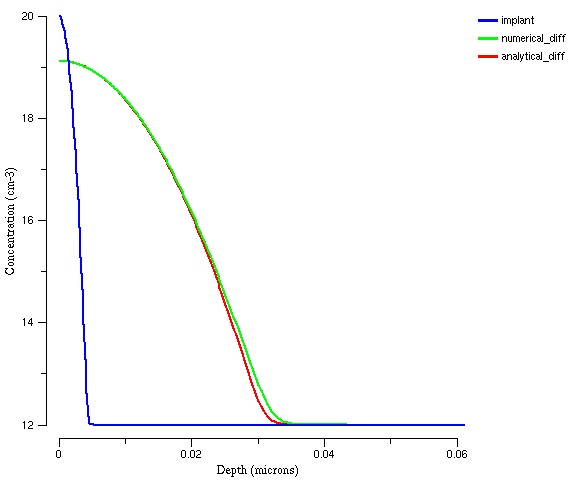
The final plot is shown below. The numerical method slightly over-predicts the diffusion distance. | |||
[[Image:diff.jpg]] | [[Image:diff.jpg]] | ||
Latest revision as of 20:09, 19 August 2010
This is an example that uses the "diffuse" command to predict diffusion of an implant profile. The numerical solution is compared to an analytical solution.
Establish diffusion parameters and solution variable "Test"
math diffuse dim=1 umf none col !scale solution add name=Test solve !negative
pdbSetDouble Silicon Test Abs.Error 1.0e-8 pdbSetDouble Silicon Test Rel.Error 1.0e-2
Establish the grid and assign a material
line x loc=0.0 spac=0.0002 tag=Top line x loc=0.2 spac=0.001 tag=Bottom region silicon xlo=Top xhi=Bottom init
Gaussian implant profile
#fill in file name for as-implanted profile here sel z=1.0e20*exp(-(x/0.001)*(x/0.001))+1.0e12 name=Test
#plot the the as-implanted profile sel z=log10(Test) plot.1d max=0.06 min=[list 0.0 12.0] label=implant
#value for diffusivity goes here term name = TestDiff add silicon eqn = "4.0e-17"
#solid-solubility limit stuff term name = TestActive add silicon eqn = "(Test>1.0e20)?1.0e20:Test"
Establish the physics
#simple Fick's law based diffusion equation pdbSetString Silicon Test Equation "ddt(Test) - TestDiff * grad(TestActive)"
Use "difuse" command to solve
diffuse time = 60.0 temp=550 init=1.0e-1
#plot the the final profile sel z=log10(Test) plot.1d !cle max=0.06 min=0.0 label=numerical_diff
Use the "FindDose" command to intigrate the total implant dose and save as "dose"
sel z=Test set dose [FindDose]
#analytical model for drive-in diffusion sel z = ($dose*1.0e4)/(sqrt(3.14*4.0e-17*1.0e8*60.0*60.0))*exp(-(x*x)/(4*4.0e-17*1.0e8*60.0*60.0))+1.0e12 name=model sel z=log10(model) plot.1d !cle max=0.06 min=0.0 label=analytical_diff
The final plot is shown below. The numerical method slightly over-predicts the diffusion distance.