PdbCommands
Paramater Data Base (pdb) Commands
Paramater Data Base (pdb) Commands - commands used to control the parameters
Synopsis
The parameter data base allows the user to configure and set various control parameters in major sections of the code. The data base is used for the diffusion, oxidation, and grid sections. There is planned expansion for other sections of the code.
All of the pdb structures are implemented in tcl. FLOOPS accesses parameters with calls back to the tcl parser to obtain values. Consequently, many of the tcl commands can be used to help manage the data base. Scripts can be written to help manage packages of parameters.
Description
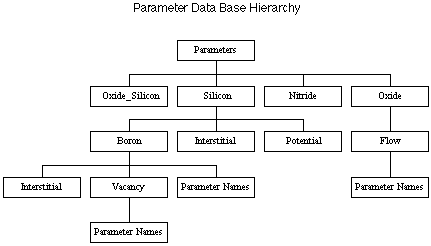
Parameter Data Base (pdb) Hierarchy
This section describes the parameter hierarchy in detail. Parameters are stored in a tree like data structure, with leaves being values. Parameters are nested in a consistent way from materials to solution values.
The pdb hierarchy is defined as a tree. Each parameter access command contains a list of keywords that define the location of the parameter in the tree. These are ordered from top down in the parameter list. The first level is material, and cna include interface material names. The second level are solution variables - boron, interstitials, clusters, flow, and oxidant. The next level below are usually parameters themselves. Some dopants have an additional nested level for the defect they diffuse with. Figure 1 illustrates part of the hierarchy.
The "Parameter Names" boxes are the locations in the leaves of the tree. In these locations, the actual name value pairs are stored. For example, the pdb hierarchy {Silicon Boron Interstitial D0} refers to the neutral diffusivity of boron when diffusing with interstitials. The pdb hierarchy {Oxide Flow Viscosity} refers to the low stress viscosity of the oxide. The [pdbBrowser.html browser] provides an excellent way to explore the hierarchy.
The hierarchy is implemented as a file system for permanent storage. Each level of the heirarchy is a directory until the lowest level where files contain the variable names and values. Each level of the hierarchy can have information tagged on it, and these values are stored in the Info file. The Info files also contain aliases for the names of the underlying levels.
Once read, the hierarchy is stored in tcl using the array features of the language. The top level is stored in the Params variable. Each of that array is a material name and the name of an array variable that stores it values.
Parameter Data Base (pdb) Commands
This describes the tcl commands and arguments that compromise the parameter access methodes. They allow general access to set and get operations, as well as bookkeeping operations. Access to the context information is also available.
In all cases, the attached commands use the {pdb Hierarchy} string as a place holder for the argument list that indicates the position of the parameter. These strings should be replaced with the correct hierarchy. Each of the commands with the {pdb Hierarchy} takes an adjustable number of arguments. They do not take a list and an argument.
Management Commands
- pdbClear
- This empties the currently stored parameters. New parameter accesses will be forced to re-read the files. This command is useful when testing new parameter values in the file structure to make sure they are entered correctly.
Parameter Access
- pdbSetDouble {pdb Hierarchy} Value
- Set a double precision variable in the hierarchy to Value. The pdb Hierarchy should start with a material and work down the tree to a leaf with type double. Value must be a term that can be passed to the expr command and evaluate to a double precision number.
- pdbSetBoolean {pdb Hierarchy} Value
- Set a boolean variable in the hierarchy to Value. The pdb Hierarchy should start with a material and work down the tree to a leaf with type boolean. Value must be a term that can be passed to the expr command and evaluate to a boolean.
- pdbSetFile {pdb Hierarchy} Value
- Set a file variable in the hierarchy to Value. The pdb Hierarchy should start with a material and work down the tree to a leaf with type file. Value must be a string that corresponds to a file.
- pdbSetSwitch {pdb Hierarchy} Value
- Set a switch variable in the hierarchy to Value. The pdb Hierarchy should start with a material and work down the tree to a leaf with type switch. Value must be a string that corresponds to one of the options available in the switch. This is a radio button style entry.
- pdbSetString {pdb Hierarchy} Value
- Set a string variable in the hierarchy to Value. The pdb Hierarchy should start with a material and work down the tree to a leaf with type string. Value must be a string.
- pdbIsAvailable {pdb Hierarchy}
- This returns a boolean. If the entry in the hierarchy is defined, the function returns true.
- pdbGetDouble {pdb Hierarchy}
- Return the double precision value corresponding to the given entry in the hierarchy.
- pdbDelayDouble {pdb Hierarchy}
- Return the double precision expression corresponding to the given entry in the hierarchy. The expression can be evaluated later - particularly useful for [../alagator.html alagator] scripts.
- pdbGetBoolean {pdb Hierarchy}
- Return the boolean value corresponding to the given entry in the hierarchy.
- pdbGetFile {pdb Hierarchy}
- Return the file name corresponding to the given entry in the hierarchy.
- pdbGetSwitch {pdb Hierarchy}
- Return the switch value corresponding to the given entry in the hierarchy. This is returned as an integer index into the list of values.
- pdbGetString {pdb Hierarchy}
- Return the string corresponding to the given entry in the hierarchy.
Examples
- pdbGetDouble Silicon Boron Interstitial D0
- Return the current value of the boron interstitial neutral diffusivity. This will depend on the last simulated temperature.
- pdbSetDouble Silicon Boron D0 {[Arrhenius 1.0 3.6]}
- Set the value of the boron interstitial neutral diffusivity. The value will depend on the temperature, using an Arrhenius relationship.
- pdbSetDouble Oxide Grid perp.add.dist 0.02e-4
- Set the desired grid spacing in oxide to 20nm.
- pdbGetSwitch Silicon Boron DiffModel
- Return the currently set diffusion model.
Bugs and Missing Features
It would be nice if the switch statement was a little easier to use. You can't set it with an integer, and it will return only integers. It needs two modes.
See Also
Individual commands that use parameters.